Holliday Junctions and Resolvases
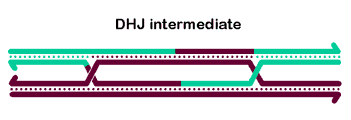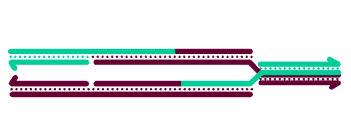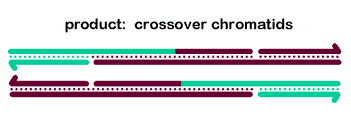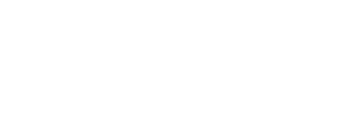
Most models for crossover formation include one or more Holliday junctions - four-stranded structures involving a partner
swap between two duplex DNA molecules. In these models, crossovers are actually made by 'resolving' the HJs through cleavage of two strands
with the same polarity; enzymes that do this are called resolvases. Several nuclear enzymes have been shown to cleave HJs in vitro, including
Mus81-Mms4/Eme1, GEN1/Yen1, and SLX4-SLX1. These are described below, along with what
we believe is a fourth resolvse - Mei-9.com.

S. pombe Mus81-Eme1 was the first candidate nuclear resolvase (Boddy 2001). This enzyme actually cuts nicked Holliday junctions and replication fork structures better than HJs (Gaillard 2003). When it cuts HJs, the ends cannot be ligated back together like canonical resolvases - hence, our choice of a chainsaw to represent this enzyme.
Drosophila Mus81-Mms4 does not have any apparent function in generating meiotic crossovers. Indeed, flies don't seem to care whether they have this enzyme or not, as Mus81 mutants are, if anything, more prolific than wild-type flies...unless the Blm helicase is also absent - then they're quite dead.
K. Trowbridge, K.S. McKim, S.J. Brill, and J. Sekelsky (2007) Synthetic lethality of Drosophila in the absence of the Mus81 endonuclease and Blm helicase is associated with elevated apoptosis. Genetics 176: 1993-2001.

Human GEN1 and S. cerevisiae Yen1 are true resolvases in vitro (Ip 2008),
cut HJs with high specificity and with precision (as can be predicted from the structure shown here). In vivo, however, they seem to play second fiddle to
Mus81-Mms4/Eme1 (Blanco 2010, Tay 2010,
Ho 2010). S. pombe doesn't even have a GEN1/Yen1 ortholog.
In Drosophila this relationship is reversed: Gen is more important than Mus81-Mms4. Gen mutants have severe hypersensitivities to DNA damaging agents
(unlike Mus81 mutants, which are only moderately sensitive to a limited number of agents) and mutants that lack both GEN and the BLM helicase
die very early in development (mutants lacking Mus81 and Blm die very late).
Although GEN1 has been described primarily as a Holliday junction resolvase, we showed, in collaboration with Dorothy Erie in UNC Chemistry, that both Drosophila Gen and human GEN1 cleave 5' flap substrates far more rapidly than they cleave HJs. This raises the question of what the relative importance of these activities in vivo.
S.L. Andersen, H.K. Kuo, D. Savukoski, M.H. Brodsky, and J. Sekelsky (2011) Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genetics 7: e1002315.
S.P. Bellendir*, D.J. Rognstad*, L.P. Morris, G. Zapotoczny, W.G. Walton, M.R. Redinbo, D.A. Ramsden, J. Sekelsky*, D.A. Erie* (2017) Substrate preference of Gen endonucleases highlights the importance of branched structures as DNA damage repair intermediates. Nucleic Acids Res. 45:5333-5348.

SLX4/Mus312 is the Swiss army knife of nuclease scaffolding proteins. In vertebrates, can interact with Mus81-EME1, SLX1, and XPF-Ercc1 simultaneously - a hexameric trinuclease complex (Wyatt 2017). Drosophila mus312 was first described in 1981 (Boyd 1981, Green 1981) (decades before the gene was discovered in any other organism). We identifed the Mus312 protein as an interactor with the nuclease Mei-9 and showed that this interaction is essential for generating meiotic crossovers. We later found that, as with the yeast and vertebrate orthologs, Mus312 interacts with the Slx1 nuclease. Mus312-Slx1 appears to be important in inter-strand crosslink repair, and is essential when Blm helicase is absent. Studies of this lethality has suggested functions for both the resolvase and the helicase in replication fork repair.
Ö. Yildiz, S. Majumder, B.C. Kramer, and J. Sekelsky (2002) Drosophila MUS312 protein interacts physically with the nucleotide excision repair endonuclease Mei-9 to generate meiotic crossovers. Molecular Cell 10: 1503-1509.
S.L. Andersen, D.T. Bergstralh, K.P. Kohl, J.R. LaRocque, C.B. Moore, and J. Sekelsky (2009) Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Molecular Cell 35: 128-135.
S.L. Andersen, H.K. Kuo, D. Savukoski, M.H. Brodsky, and J. Sekelsky (2011) Three structure-selective
endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genetics 7: e1002315.

Mei-9 and Ercc1 encode the Drosophila orthologs of S. cerevisiae Rad1p and Rad10p, and mammalian XPF/ERCC4 and Ercc1. The heterodimer is an endonuclease that recognizes and cleaves DNA strands that transit from 5' double-stranded to 3' single-stranded, such as bubbles generated during nucleotide excision repair (NER) (Bardwell 1994, Park 1995). Mei-9 and Ercc1 mutants are hypersensitive to many types of DNA damaging agents, including ionizing radiation, ultraviolet radiation, and interstrand crosslinking agents. These hypersensitivities underlie some of the pathways in which the endonuclease functions, including NER and ICL repair. Interestingly, Mei-9 and Ercc1 mutants also have defects in meiotic recombination, including a 90% decrease in crossovers. We hypothesized that Mei-9-Ercc1 cuts Holliday junctions to generate crossovers, as shown in the animation at the top of this page.
In one model, in the absence of Mei-9 the double-HJ undergoes dissolution, as shown at right. Because there are no nicks made during dissolution, mismatch repair does not occur, leading to post-meiotic segregation (PMS) of markers, seen as mosaic progeny. Carpenter (1982) observed PMS in Mei-9 mutants. We mapped PMS tracts and showed that they exhibit the "trans" arrangement predicted by the model.
S.J. Radford, S. McMahan, H. Blanton, and J. Sekelsky (2007) Heteroduplex DNA in meiotic recombination in Drosophila Mei-9 mutants Genetics 176: 63-72.
As described above, a combination of genetic and protein interaction studies identified Mus312 as an interactor. We call this complex, possibly including the OB-fold protein HDM (Joyce 2009) Mei-9.com (Mei-9 complex). We've made several attempts to express and purify Mei-9.com, so far without success.
Ö. Yildiz, S. Majumder, B.C. Kramer, and J. Sekelsky (2002) Drosophila MUS312 protein interacts physically with the nucleotide excision repair endonuclease Mei-9 to generate meiotic crossovers. Molecular Cell 10: 1503-1509.

