Mitotic recombination was first described by Curt Stern, working with Drosophila, in a classic paper published in 1936 (Stern 1936). Although meiotic recombination is frequent and programmed, mitotic recombination is generally rare and abnormal. Mitotic recombination events are thought to have their origins in cellular problems, such as replication fork blockages or double-strand DNA breaks (DSBs) resulting from oxidative stress.
In cells proliferating mitotically, the strategy for repairing a DSB involves a series of decsion points:
Resect the ends? Canonical non-homologous end-joining (NHEJ) can join ends that are blunt or, with some processing, that have short overhangs or damaged bases. Alternatively, the 5' ends can be resected to leave long 3'-ended single-stranded sections. After resection, NHEJ is no longer an option, making homology-directed repair (HDR) the preferred strategy to promote fidelity.
Anneal complementary sequences? If there are complementary single-stranded sequences, they can be annealed. Overhangs are trimmed, gaps are filled, ligation occurs, and voila! - repair is done. In most cases, complementary sequences will not be available until after the next two steps. We put this possibility here to accommodate the special case of when a DSB is generated between repeated sequences.
Search for a homologous duplex template? If there are no complementary ssDNA sequences to anneal, the most common course is the assemble a Rad51 filament on one or both ssDNA ends and do a homology search. Rad51 and other proteins then catalyze strand exchange, forming a D-loop (see below) and synthesis ensues.
An alternative is to complete repair by DNA polymerase theta-mediated end joining (TMEJ, also known as alt-EJ or MMEJ).Disassemble D-loop? After some amount of synthesis, the D-loop can be diassembled by a helicase (and probably a topoisomerase). Now that one 3' end has been extended, it should be able to anneal to the other resected end of the DSB - go to step 2. For gaps (e.g., after TE excision) there may be several cycles through steps 2-4.
If the D-loop is not disassembled, the 2nd resected end can be annealed to the displaced single strand. Additional synthesis and ligation generated a double Holliday-junction (dHJ). Please see our page on meiotic recombination for a discussion of the dHJ model.
We beleive the most common pathway for HDR in mitotic cells is steps 1-3-4-2, which together describe the synthesis-dependent strand annealing (SDSA) model. A gif animation of SDSA is shown below.
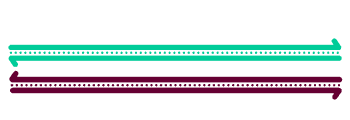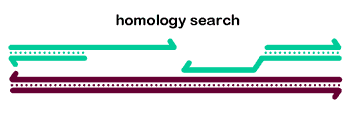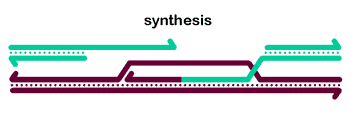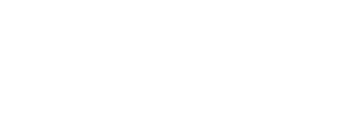
We first showed that Drosophila Blm helicase has an important role in promoting SDSA. We also provided evidence for multiple cycles of strand invasion, synthesis, and dissociation during repair of a gap.
H.K. Kuo, S.McMahan, C.M. Rota, K.P. Kohl, and J. Sekelsky (2014) Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics 198: 935-945.
M. McVey, M.D. Adams, E. Staeva-Vieira, and J. Sekelsky (2004) Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699-705.
M. McVey, J. LaRocque, M.D. Adams, and J. Sekelsky (2004) Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694-15699.
M.D. Adams, M. McVey, and J. Sekelsky (2003) Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265-267.
A key feature of SDSA is that it prevents mitotic crossovers, which can lead to loss of heterozygosity or chromosome rearrangment (in the case of non-allelic repair templates). We showed the Drosophila Blm and Fancm mutants have elevated crossovers, and we've sequenced some of these to gain insight into sources and mechanisms of repair.
M.C. LaFave., S.L. Andersen, E.P. Stoffregen, J.K. Holsclaw, K.P. Kohl, L.J. Overton, and J. Sekelsky (2014) Sources and structures of mitotic crossovers that arise when BLM helicase is absent in Drosophila. Genetics 196: 107-118.
H.K. Kuo, S.McMahan, C.M. Rota, K.P. Kohl, and J. Sekelsky (2014) Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics 198: 935-945.
M. McVey, S. Andersen, Y Broze, and J. Sekelsky (2007) Multiple functions of the Drosophila Blm helicase in maintaining genome stability. Genetics 176: 1979-1992.

