The challenge of meiosis is to accurately segregate homologous chromosomes from one another to generate haploid gametes for sexual reproduction. Although many strategies have evolved to do this, the most common one relies on the formation of crossovers between homologous chromosomes. Sites of crossing over mature into chiasmata - physical connections between homologous chromosomes. These connections facilitate stable alignment on the meiotic spindle, with homologous kinetochores oriented toward opposite spindle poles.
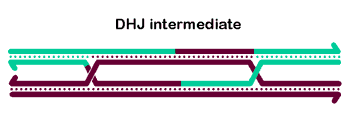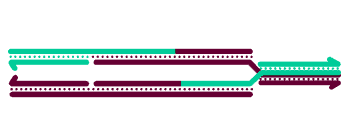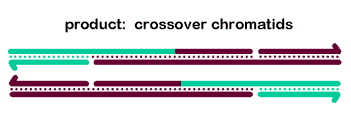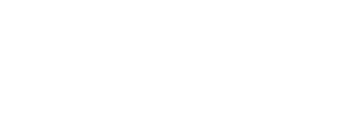
Most current models for meiotic crossing over are based on the dHJ model described below. It is likely that many of the details of the model depicted are incorrect, but the general pathway is probably similar to one described, at least in some organisms.
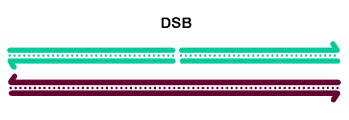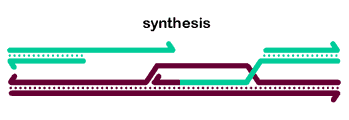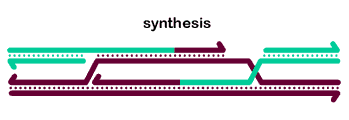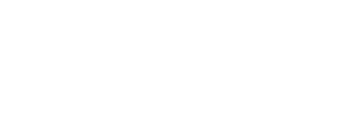
The basic steps in the canonical model to generate a double-Holliday junction (dHJ) intermediate are:
Initiation involves formation of a double-strand break by Spo11 (MEI-W68 in Drosophila) and friends.
The 5' ends undergo resection, leaving 3'-ended single-stranded tails.
One tail commits Rad51-mediated strand invasion.
Repair DNA synthesis extends the invading strand.
2nd end capture occurs when the D-loop displaced by synthesis anneals to the other end of the break.
Ligation of the free ends results in formation of a double Holliday junction (dHJ) intermediate.
To generate crossover chromatids, the dHJ must undergo resolution. The most commonly hypothesized resolution mechanism involves nicking at each junction by a resolvase, swapping of the ends, and ligation. At each Holliday junction, two strands with the same polarity are nicked. A crossover occurs when different strands are nicked at each HJ, so that each of the four strands receives a single nick. The figure at right is a two-dimensional projection, so we've drawn it as if the right end rotates around to allow ligation to the other strands' former partners.
Meiotic recombination can also produce non-crossover chromatids, which can be detected if gene conversion occurs. Non-crossovers might arise from the same resolution process as described above, if the same two strands as nicked at each junction (not shown), but studies in yeast suggest that most meiotic non-crossovers arise through SDSA.
Our interests in meiotic recombination focus on:
Crossover patterning: How is the decision to produce a crossover versus a non-crossover determined, and how is it carried out? We are addressing this by examining the function of the mei-MCM complex and Bloom syndrome helicase. We are also directly studying the mechanism of the centromere effect on recombination.
T. Hatkevich,* K.P. Kohl, S. McMahan, M.A. Hartmann, A.M. Williams, and J. Sekelsky (2017) Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Current Biology.
.K.P. Kohl, C.D. Jones, and J. Sekelsky (2012) Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363-1365.
H. Blanton, S.J. Radford, H. Kearney, S. McMahan, J. Ibrahim, and J. Sekelsky (2005) REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genetics 1: e40.
dHJ resolution: Little is known about meiotic HJ resolvases. We believe that a complex comprising the MEI-9-ERCC1 endonuclease, the nuclease scaffolding protein MUS312, and the OB fold protein HDM is the primary resolvase in Drosophila meiosis. We are analyzing this complex genetically and biochemically. We have also knocked out all mismatch repair so we can analyze heteroduplex DNA patterns to give us insights into what the pre-crossover intermediates really look like.
K.N. Crown, S. McMahan, and J. Sekelsky (2014) Eliminating both canonical and short-patch mismatch repair in Drosophila suggests a new meiotic recombination model. PLoS Genetics 10: e1004583.
Ö. Yildiz, S. Majumder, B.C. Kramer, and J. Sekelsky (2002) Drosophila MUS312 protein interacts physically with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Molecular Cell 10: 1503-1509.
S.J. Radford, S. McMahan, H. Blanton, and J. Sekelsky (2007) Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants Genetics 176: 63-72.

